about
This website is a tool designed to explore a set of cancer genomes collected and analyzed by the Samsung Medical Center and The Rabadan Lab at Columbia University Medical Center.
This website is designed to take you, the user, through various "paths" of our dataset. Each feature you select serves as a filter to reduce the number of samples. Once you are satisfied with the subset of samples, you can see how a particular drug affected that subset.
Suppose you have analyzed a cancer patient's genome in your own lab and are curious to know how a particular drug might affect your patient. Our tool is designed to showcase, in a user-friendly fashion, how the drug in question affected the patients in our study with a similar genetic profile (though the statistical power is limited).
research
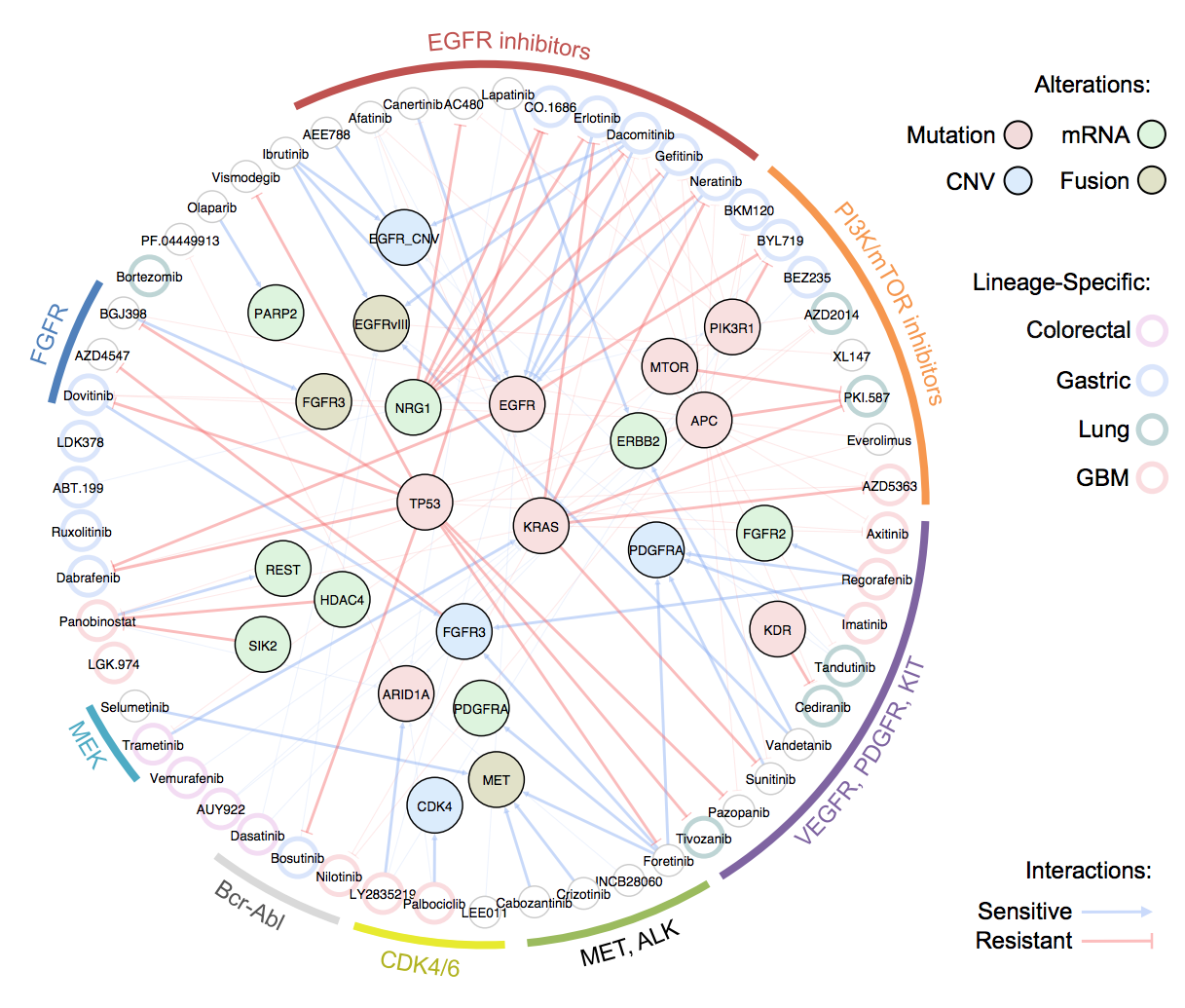
A fundamental tenet of precision oncology is that genomic and molecular tumor profiling enable identification of the most effective drugs tailored to the patient. However, predicting successful therapies on the basis of computational approaches alone may be challenging because the drug response could vary according to the genomic complexity and heterogeneity of the tumor.
An integrated approach consisting of genomic analysis of the patient tumor, direct measures of tumor growth in response to drugs using patient tumor derived cells (PDCs) would represent a next step toward implementing precision oncology.
The aims of our research are to analyze pharmacogenomic interactions that can not only evaluate the previously known genomic targets for a particular drug, but also explore the unknown mechanisms of drug sensitivity or resistance.
Here we have constructed therapeutic landscape of 462 genomically annotated PDCs across 14 cancer types including, including malignant gliomas, brain metastasis, gastric, colorectal, lung, breast, ovary cancers, renal cell carcinoma and sarcomas, and 60 anti-cancer drugs margeting major oncogenic signaling pathways
We demonstrated several valuable pharmacogenomic interactions including 1) genotype-specific drug responses in selected cancer types. 2) frequency of alterations in a selected gene according to the cancer types, and 3) cancer type specific drug sensitivities.
This study is a result of international collaboration between Samsung Medical center in South Korea (Patient-derived cell culture, sequencing and drug screening) and Columbia University in USA (Analysis of Pharmacogenomic interactions and portal-site development). This work was supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea. This work has been funded by NIH grants (R01 CA185486, R01 CA179044, U54 CA193313 and U54 209997) and NSF/SU2C/V-Foundation Ideas Lab Multidisciplinary Team (PHY-1545805).
tutorial
The Cancer-Drug Explorer website is designed to help the user navigate the dataset in two main ways:
1) The 'genomic' tab allows the user to filter the dataset by any subset of cancers, genes, drugs, or features. Selecting an option in one of three 'branches' will take the user through a step by step process of filtering the dataset.
2) The 'transcriptomic' tab allows the user to filter the dataset based on gene expression data.
The user can switch between the 'explore' and 'expression' tabs by clicking the tabs at the top. Switching tabs will not reset the visualizations in the tab you are switching from.
The user can navigate down through each branch either by selecting an option from the toolbar on the left, or by clicking values they are interested in on the visualizations on the right.
The user can also navigate up through each branch by either clicking the ← button to move up one step in the branch, or by clicking 'clear' or 'reset' below a selection to jump back to that option.
We also provide a download button located at the bottom of the page for most sections as an easy way for the user to obtain the data for the visualization on display.

